В последние годы для эпидемиологической характеристики штаммов патогенов, наряду с классическими методами, широкое применение нашли методы генетического анализа, позволяющие не только обнаружить искомый патоген, но и изучить его на молекулярно-генетическом уровне, в том числе ретроспективно.
Модернизация системы мониторинга за возбудителем бруцеллеза неотъемлемо связана с необходимостью внедрения современных (оптимизированных) алгоритмов молекулярно-эпидемиологического анализа штаммов бруцелл, циркулирующих на отдельных территориях, и последующей геномной паспортизации патологических биологических агентов. Разработка алгоритмов идентификации изолятов, ассоциированных с топографией выделения, имеет важнейшее значение для проведения эпидемиологических расследований единичных случаев и вспышек бруцеллеза, особенно на эпизоотически условно благополучных территориях.
Среди молекулярно-генетических методов, используемых для характеристики бруцелл, широкое применение получила полимеразная цепная реакция (ПЦР) с электрофоретической детекцией результатов, а в последние годы – с учетом результатов в режиме реального времени [1, 2]. Основной целью данной методики является дифференциация бруцелл до вида, а в некоторых случаях – до биовара.
В качестве эффективных подходов для генетического типирования применяются методы MLST (multilocus sequence typing) и SNP (single nucleotide polymorphism ) [3], однако использование данных методик является длительным, трудоемким и дорогостоящим и требует участия высококвалифицированного персонала.
В настоящее время для генетического типирования бруцелл используют метод MLVA, представленный несколькими схемами, содержащими от 8 до 80 VNTR-локусов с размерами повторов от 5 до 339 п.н. [4]. Однако данный метод имеет ряд существенных недостатков. Например, для детекции локусов с длиной мотива менее 10 п.н. необходимо использовать сложные и трудоемкие методики определения молекулярных размеров амплифицированного фрагмента с применением капиллярных анализаторов. В ряде случаев MLVA проводят с использованием зондов с флуоресцентной меткой, что усложняет и удорожает процедуру синтеза праймеров. Для учета результатов необходим высокотехнологичный дорогостоящий секвенатор и импортные расходные материалы.
Одним из простых и информативных методов типирования является выявление INDEL-маркеров, представляющих собой вставки-делеции (insertions/deletions) нескольких нуклеотидов. Изучение полиморфизма INDEL-генов широко используется при молекулярном типировании микроорганизмов [5].
Цель исследования – INDEL-типирование по 10 локусам штаммов B. melitensis, выделенных в разных географических регионах мира, в том числе на территории юга европейской части Российской Федерации.
Материалы и методы
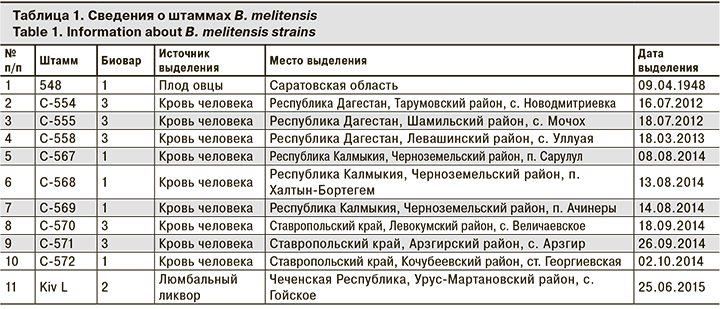
В работе использовали 11 штаммов B. melitensis, выделенных в период с 1948 по 2015 г., из коллекции патогенных микроорганизмов ФКУЗ «Ставропольский противочумный институт» Роспотребнадзора (табл. 1).
Обеззараживание и подготовку проб проводили согласно СП 1.3.3118-13 «Безопасность работы с микроорганизмами I–II групп патогенности (опасности)» и МУ 1.3.2569-09 «Организация работы лабораторий, использующих методы амплификации нуклеиновых кислот при работе с материалом, содержащим микроорганизмы I–IV групп патогенности».
Для генетического типирования были выбраны 10 INDEL-локусов, для амплификации которых были сконструированы специфичные праймеры (табл. 2).

ДНК выделяли с помощью комплекта реагентов «ДНК-сорб-B» (ЦНИИ эпидемиологии Роспотребнадзора, Россия) в соответствии с инструкцией производителя.
Реакционную смесь объемом 20 мкл готовили из расчета 5 мкл MgCl2-буфера, 2,5 мкл смеси дезоксинуклеотидтрифосфатов, по 0,2 мкл каждого праймера, 0,5 мкл ДНК-полимеразы, 1,6 мкл РНК-элюента и 10 мкл ДНК-матрицы.
Микропробирки помещали в термоциклер и проводили амплификацию по схеме: денатурация 95 °C – 15 мин; 30 циклов (денатурация 95 °C – 15 с, отжиг 56 °C – 20 с, синтез 72 °C – 15 с); 72 °C – 5 мин.
Результаты амплификации учитывали методом электрофореза в 2% агарозном геле при напряженности электрического поля 10 В/см. Анализ результатов проводили по наличию специфических полос амплифицированной ДНК на электрофореграмме, затем определяли размеры полученных ампликонов по всем локусам для каждого исследуемого штамма, сравнивая их с маркером молекулярного размера.
Генотипирование in silico проводили, включая геномные последовательности 48 штаммов B. melitensis, находящиеся в открытом доступе в международной базе данных GenBank (https://ftp.ncbi.nlm.nih.gov/ genomes/genbank/bacteria/Brucella_melitensis) (табл. 3).

Для численной оценки дискриминирующей способности метода генетического типирования использовали индекс Хантера–Гастона (HGDI).
Статистическую обработку данных проводили в среде языка R [6] с использованием пакетов «tidyverse» [7] и «pheatmaply» [8]. Дендрограммы строили на основе метода полной связи (Complete-linkage clustering).
Результаты
Штаммы B. melitensis, используемые в исследовании, представлены описанными нами ранее 5 основными wgSNP-генотипами (I–V), которые соответствуют потенциальному географическому происхождению изолятов [9]. Штаммы, выделенные в странах Средиземноморья, были идентифицированы как генотип I, азиатские штаммы отнесены к генотипу II, генотип III представлен штаммами африканского происхождения. Генотипы IV и V отнесены к европейской и американской линиям соответственно.
Полученные в работе данные INDEL-типирования позволили установить, что исследуемые штаммы B. melitensis представлены 5 INDEL-генотипами, обозначенными буквами А, B, C, D и E (рисунок, см. на вклейке).
Сопоставление данных INDEL-типирования штаммов возбудителя бруцеллеза и wgSNP-анализа – основного метода для определения вероятного географического региона происхождения микроорганизма, дало возможность выявить очевидную взаимосвязь INDEL-генотипа штамма с его полногеномным SNP-профилем.
Генотип А (98, 91, 76, 86, 84, 61, 80, 72, 76, 78) включал штаммы, выделенные преимущественно в странах Средиземноморья (Египет, Италия). Все штаммы генотипа А соответствовали wgSNP-генотипу I, диверсификация которого могла иметь место около 6,5 тыс. лет назад в эпоху неолита.
Штаммы второго, наиболее представительного генотипа В (98, 91, 88, 86, 84, 61, 80, 72, 76, 78) имели самое широкое географическое распространение, включая большую часть территории Евразийского континента. Примечательно, что к указанному INDEL-генотипу принадлежали все исследованные штаммы B. melitensis, выделенные на территории Российской Федерации. Генотипы А и В различались размером лишь 1 из 10 INDEL-локусов – RS12095 (76 и 88 п.н. соответственно).
Согласно ранее опубликованным данным полногеномного анализа [9], штаммы генотипа В соответствовали генетической линии II, которая сформировалась около VII в. Весьма вероятно, что многочисленные представители этой генетической линии, объединяющей 9 подгенотипов, имеют общего предка средиземноморского происхождения. Можно с большой уверенностью предположить, что изоляты B. melitensis генотипа В II произошли от так называемой восточно-средиземноморской линии, штаммы которой наиболее распространены в неблагополучных по бруцеллезу странах Ближнего Востока и Центральной Азии, имеющих длительную историю активного бартера и внешней торговли жвачными животными (овцами и козами), особенно в период экономического процветания региона – функционирования экономического пояса «Шелкового пути» [10].
Генотип С (77, 67, 76, 74, 84, 61, 80, 72, 76, 78) был представлен двумя штаммами: UK22/04 (Великобритания, 2004) и UK24/06 (Нигерия, 2006). Наиболее генетически близким к нему генотипом D (77, 67, 76, 74, 84, 61, 80, 67, 76, 78) обладали штаммы, выделенные в Африке (Сомали, Кувейт). Генотипы С и D различались аллельным вариантом INDEL-локуса RS16525 (72 и 67 п.н. соответственно). По глобальной классификации на основе wgSNP, штаммы генотипов С и D были отнесены к «африканскому» генотипу III, дивергенция которого могла произойти около 3,5 тыс. лет назад.
Генотип E (98, 67, 76, 74, 71, 65, 89, 72, 67, 61) имел смешанный состав относительно географического происхождения штаммов. Этот генотип был определен для бруцелл, выделенных в Европе (Норвегия, Португалия, Мальта), Америке (Аргентина) и 1 штамма в Азии (Индия). Следует отметить, что все европейские изоляты кластера относились к сравнительно редкому wgSNP-генотипу IV, отделившемуся от предковой ветви генотипа III и занесенному на территорию Европы, вероятно, с побережья Северной Африки. Очевидная малочисленность известных штаммов этой генетической линии может быть связана с тем, что страны Северной и Западной Европы добились практически полной ликвидации бруцеллеза сельскохозяйственных животных.
Штаммы генотипа E, выделенные в Аргентине и Индии, имели «американский» wgSNP-генотип V, сформировавшийся около X в.
Обсуждение
Стабильность INDEL-генотипа штаммов B. melitensis определенного географического происхождения, выделенных в разные годы, свидетельствует о высокой степени консервативности выбранных локусов.
Значение индекса HGDI, рассчитанное на основании анализа аллельного полиморфизма 10 INDEL-локусов, составило 0,92, что демонстрирует высокую разрешающую способность разработанного метода.
Важными преимуществами представленного метода INDEL-типирования B. melitensis по сравнению с полногеномным SNP-анализом являются экономическая эффективность и простота исполнения при наличии стандартного ПЦР-оборудования. В то же время разрешающая способность wgSNP-типирования, несомненно, выше и позволяет установить, помимо принадлежности изолята к одной из 5 основных генетических линий, конкретный подгенотип микроорганизма, что имеет особое значение при анализе штаммов наиболее распространенного «евразийского» генотипа II, подгенотипы которого (IIh и IIi) характерны для территории Российской Федерации.
На разработанный метод генетического типирования получен патент Российской Федерации на изобретение «Способ генетического INDEL-типирования штаммов Brucella melitensis» (RU 2732425 C1) [11].
Заключение
Таким образом, предложенный метод INDEL-типирования позволяет с высокой разрешающей способностью дифференцировать штаммы B. melitensis в соответствии с основными генетическими линиями глобальной популяции возбудителя бруцеллеза. Установлено, что штаммы B. melitensis, циркулирующие на территории Российской Федерации, относятся к наиболее распространенному INDEL-генотипу В.
Результаты исследования выявили высокую степень сходства INDEL-генотипов штаммов B. melitensis, циркулирующих на одной территории, что дает возможность рассматривать разработанный метод INDEL-типирования в качестве дополнительного инструмента для определения происхождения отдельных изолятов B. melitensis при эпидемиологических расследованиях. Вместе с тем применение INDEL-типирования в комплексе с другими методами молекулярной эпидемиологии позволит конструировать модели пространственно-временной диссеминации возбудителя бруцеллеза на изучаемой территории.


